Change Log¶
This page lists changes for the Beacon+ implementation of the "Beacon" genomics API, as well as related updates for the Progenetix front-end.
2025-04-16: Reverting to pgx:icdot-55003 for PDAC tumors¶
In contrast to the original ICD-O system which limits the 8500/3 code to breast
as locus/organ, newer use (e.g. WHO Blue Books, TCGA, NCI...) allow/require the use
of 8500/3 for pancreatic adenocarcinomas (PDAC). Interestingly the 2025 SEER
mappings allow both 8140/3 (Adenocarcinoma, NOS) and 8500/3 (Ductal Adenocarcinoma, NOS)
for pancreas whwereas the Blue Book only list 8500/3. So, as compromise we now
remapped all PDAC (where identified easily as such) to pgx:icdom-85003 (Ductal
Adenocarcinoma, NOS) but left the other codes as pgx:icdom-81403.
db.biosamples.updateMany({"histological_diagnosis.id":"NCIT:C9120"}, {$set:{"icdo_morphology":{"id":"pgx:icdom-85003", "label":"Infiltrating duct carcinoma, NOS"}}})
db.biosamples.updateMany({"icdo_topography.id":{$regex:/pgx:icdot-C25.\d/}, "notes":{$regex:/^.*?pancr.*?duct.*?carc.*?$/i}}, {$set:{"icdo_morphology":{"id":"pgx:icdom-85003", "label":"Infiltrating duct carcinoma, NOS"}}})
2025-03-20: Fixing PMID: => pubmed:¶
At some point in the far past identifiers.org did use PMID as PubMed CURIE prefix.
AFAWK at some point already years ago NCBI pstarted to use pubmed ... and since identifiers.org
doesn't resolve PMID anymore we're switching prefixes, too. Hope hiccups will be limited...
var dbs = ["progenetix", "refcnv", "cellz", "examplez"]
dbs.forEach(function(database) {
db = db.getSiblingDB(database)
db.collations.find({"id":{$regex:/PMID/}}).forEach(function(doc) {
nid = doc.id.replace('PMID', 'pubmed');
db.collations.updateOne({"_id": doc._id}, {$set:{"id": nid, "namespace_prefix": "pubmed", "parent_terms": [nid], "child_terms": [nid], "hierarchy_paths": [ { "order": 0, "depth": 0, "path": [ nid ] } ]}});
})
db.biosamples.find({"references.pubmed.id":{$regex:/PMID/}}).forEach(function(doc) {
nid = doc.references.pubmed.id.replace('PMID', 'pubmed');
db.biosamples.updateOne({"_id": doc._id}, {$set:{"references.pubmed.id": nid}});
})
})
use _byconServicesDB
db.publications.find({"id":{$regex:/PMID/}}).forEach(function(doc) {
nid = doc.id.replace('PMID', 'pubmed');
db.publications.updateOne({"_id": doc._id}, {$set:{"id": nid}});
})
2025-03-12: Sonme NCIT changes¶
db.biosamples.updateMany({"histological_diagnosis.id":{$in:["NCIT:C28327", "NCIT:C66951"]}},{$set:{"histological_diagnosis":{"id":"NCIT:C127907", "label":"Endocervical Adenocarcinoma, Usual-Type"}}})
db.biosamples.updateMany({"histological_diagnosis.id":{$in:["NCIT:C66953"]}},{$set:{"histological_diagnosis":{"id":"NCIT:C180869", "label":"Cervical Mucinous Adenocarcinoma, Not Otherwise Specified"}}})
db.biosamples.updateMany({"histological_diagnosis.id":{$in:["NCIT:C4462"]}},{$set:{"histological_diagnosis":{"id":"NCIT:C4614", "label":"Skin Papilloma"}}})
db.biosamples.updateMany({"histological_diagnosis.id":{$in:["NCIT:C3234"]}},{$set:{"histological_diagnosis":{"id":"NCIT:C4456", "label":"Malignant Mesothelioma"}}})
db.biosamples.updateMany({"histological_diagnosis.id":{$in:["NCIT:C4250"]}},{$set:{"histological_diagnosis":{"id":"NCIT:C6505", "label":"Atypical Lipomatous Tumor/Well Differentiated Liposarcoma"}}})
db.biosamples.updateMany({"histological_diagnosis.id":{$in:["NCIT:C3753"]}},{$set:{"histological_diagnosis":{"id":"NCIT:C6284", "label":"Brain Germinoma"}}})
db.biosamples.updateMany({"histological_diagnosis.id":{$in:["NCIT:C27892"]}},{$set:{"histological_diagnosis":{"id":"NCIT:C9385", "label":"Renal Cell Carcinoma"}}})
db.biosamples.updateMany({"histological_diagnosis.id":{$in:["NCIT:C9385"]}},{$set:{"icdo_morphology":{"id":"pgx:icdom-8312", "label":"Renal cell carcinoma, NOS"}}})
db.biosamples.updateMany({"histological_diagnosis.id":{$in:["NCIT:C128696"]}},{$set:{"histological_diagnosis":{"id":"NCIT:C4340", "label":"Peripheral T-Cell Lymphoma, Not Otherwise Specified"}}})
db.biosamples.updateMany({"histological_diagnosis.id":{$in:["NCIT:C3916"]}},{$set:{"histological_diagnosis":{"id":"NCIT:C156757", "label":"Parathyroid Gland Adenoma"}}})
db.biosamples.updateMany({"histological_diagnosis.id":{$in:["NCIT:C2996"]}, "icdo_topography.id":"pgx:icdot-C56.9"},{$set:{"histological_diagnosis":{"id":"NCIT:C8106", "label":"Ovarian Dysgerminoma"}}})
db.biosamples.updateMany({"histological_diagnosis.id":{$in:["NCIT:C2996"]}, "icdo_topography.id":"pgx:icdot-C62.9"},{$set:{"histological_diagnosis":{"id":"NCIT:C8591", "label":"Testicular Germ Cell Tumor"}}})
db.biosamples.updateMany({"histological_diagnosis.id":{$in:["NCIT:C3110"]}},{$set:{"histological_diagnosis":{"id":"NCIT:C4858", "label":"Placental Neoplasm"}}})
db.biosamples.updateMany({"icdo_morphology.id":"pgx:icdom-92313"},{$set:{"histological_diagnosis":{"id":"NCIT:C27502", "label":"Extraskeletal Myxoid Chondrosarcoma"},"icdo_topography":{"id":"pgx:icdot-C49.9", "label":"Connective, subcutaneous and other soft tissues, NOS"}}})
2025-02-11: Introducing the VRS derived type parameter for variants¶
The internal variant model was updated to include the VRS v2 type parameter. In
contrast to the VRS model we just use it in the variant's root. So far, only
Allele, CopyNumberChange and Adjacency are used.
Update procedure, which in that form (and order) might be specific for Progenetix:
db.variants.updateMany({"variant_state.id":{$regex:/^EFO/}},{$set:{"type":"CopyNumberChange"}})
db.variants.updateMany({"variant_state.id":{$regex:/^SO/}},{$set:{"type":"Allele"}})
db.variants.updateMany({"adjoined_sequences":{$exists:true}},{$set:{"type":"Adjacency"}})
db.variants.updateMany({"adjoined_sequences":{$exists:true}},{$unset:{"fusion_id":""}})
2025-02-10: Fix for bug in aggregation of individuals.sex values¶
Due to a bug in the hierarchy codes for the individuals.sex.id parameter since
switching from PATO to NCIT codes the aggregated data for "male" sex contained
all individuals with an annotated sex, i.e. both males and females.
Performed normalization steps:
- fixing of the hierarchy, now reading as
NCIT:C27993 general qualifier 0 1
NCIT:C17998 unknown 1 2
NCIT:C20197 male 1 3
NCIT:C16576 female 1 4
- reassignment of missing codes to
NCIT:C17998
Commands run in the MongoDB backend:
db.individuals.updateMany({"sex.id":""},{$set:{"sex":{"id":"NCIT:C17998", "label":"unknown"}}})
db.individuals.updateMany({"sex.id":"PATO:0020000"},{$set:{"sex":{"id":"NCIT:C17998", "label":"unknown"}}})
- reaggregation of the
individuals.sexvalues (here forprogenetix)
./housekeepers/collationsCreator.py -d progenetix --collationTypes NCITsex
./housekeepers/collationsFrequencymapsCreator.py -d progenetix --collationTypes NCITsex --limit 0
2024-11-02: Fix for missing TCGA analyses¶
During import of the TCGA non-CNV variants the "analysis" objects somehow got
lost although variants were imported correctly, leading to a missing association of
those variants w/ their samples during query result aggregation. Fixed for all
10007 samples by extracting the previously assigned analysis_id values together
with the biosample_id values from the variants and generating the missing analysis
objects. Now TCGA has working calls for "double hit" CNV + SNV events.
2024-09-10: Ongoing removal of samples from pure germline series¶
So far of the currently 145264 samples the Progenetix collection consisted of besides data from
- 112674 cancer samples (including primary tumors, recurrences and metastases)
- 5742 cancer cell line samples
- 26848 germline samples consisting of
- germline references of tumor patients
- 3201 reference profiles from the 1000 genomes project
- samples from 58 "accidental" germline series (i.e. such being analyzed in automated download & processing and then being kept for comparative purposes)
We are now in the process of creating a new resource for the germline samples which will become available in the near future under the refcnv.org address. While strengthening the focus of the Progenetix database this will provide a new general resource for germline CNV data e.g. as reference for rare disease applications.
2024-06-18 Switching ontology use for individual.sex¶
While we had previously used the PATO terms for genotypic sex (PATO:0020001
or PATO:0020002) we have now recoded those to "NCIT:C16576": "female" and "NCIT:C20197": "male"
to stay in line with the Beacon v2 documentation examples (which will probably drive implementations).
sex:
label: Genotypic Sex
infoText: |
Genotypic sex of the individual.
defaultValue: ""
isHidden: true
options:
- value: ""
label: "(no selection)"
- value: NCIT:C16576
label: female
- value: NCIT:C20197
label: male
2024-02-24 Adding analysis_operation to analyses¶
This new parameter with its (so far) values
"analysis_operation.id":"EDAM:operation_3961", "analysis_operation.label":"Copy number variation detection""analysis_operation.id":"EDAM:operation_3227", "analysis_operation.label":"Variant Calling"
... allows now the filtering of analyses based on the type of genomic profiling performed.
2023-06-12: Some schema changes...¶
Schema changes are now extensively tracked in bycon.progenetix.org
The latest changes of the database schemas involve e.g.:
- several field changes in biosamples, to align w/ main Beacon v2 default schema:
sampledTissue=>sampleOriginDetaildescription=>notestimeOfCollection.age=>collectionMoment
2022-12-22: Various API fixes & extensions¶
- the
genespansresponses contain now acytobandsparameter (i.e. the cytogenetic mapping of the gene's locus) - FIXED: the beacon response meta now has the standard
receivedRequestSummary.requestParametersinstead of beforereceivedRequestSummary.variantPars
2022-01-17: Term-specific queries¶
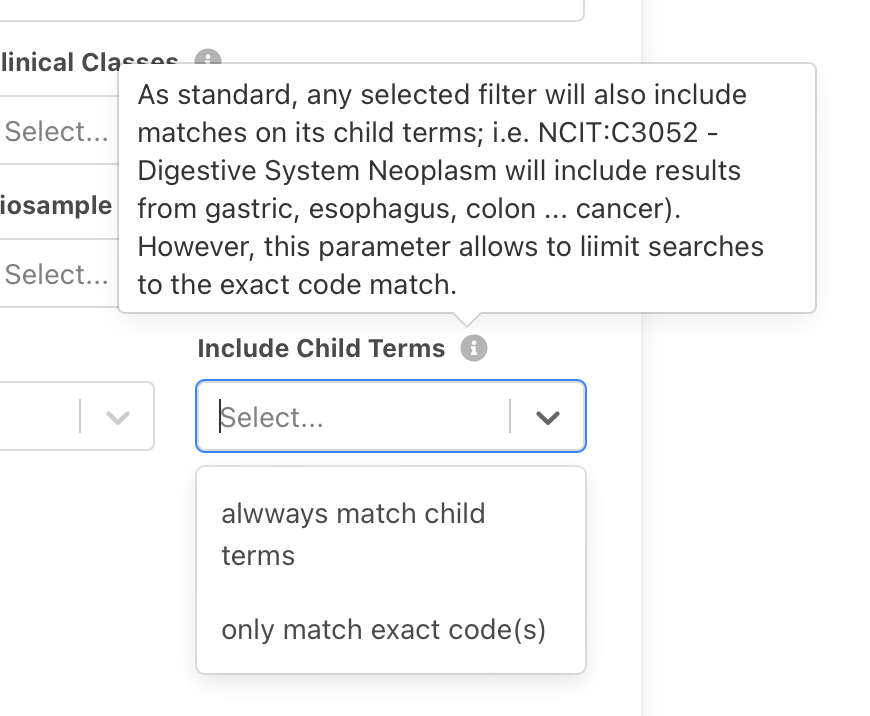 So far (and still as standard), any
selected filter will also include matches on its child terms; i.e. "NCIT:C3052 -
Digestive System Neoplasm" will include results from gastric, esophagus, colon
... cancer. Here we introduce a selector for the search panel to make use of the Beacon v2
filters
So far (and still as standard), any
selected filter will also include matches on its child terms; i.e. "NCIT:C3052 -
Digestive System Neoplasm" will include results from gastric, esophagus, colon
... cancer. Here we introduce a selector for the search panel to make use of the Beacon v2
filters includeDescendantTerms pragma, which can be set to false if one only
wants to query for the term itself and exclude any child terms from the matching.
Please be aware that this can only be applied globally and will affect all filtering terms used in a query. More information is available in the Filtering Terms documentation.
2022-01-17: Introducing variant_state classes for CNVs¶
More information can be found in the description of ontology use for CNVs.
2022-01-10: BUG FIX Frequency Maps¶
Pre-computed Progenetix CNV frequency histograms (e.g. for NCIT codes) are based
samples from all child terms; e.g. NCIT:C3262 will display an overview of all
neoplasias, although no single case has this specific code.
However, there had been a bu when under specific circumstances (code has some mapped samples and code has more samples in child terms) only the direct matches were used to compute the frequencies although the full number of samples was indicated in the plot legend. FIXED.