Progenetix News¶
Data model and plotting update
Now with "high level" support
Both the data model and the plot engine now support an separate color style for high level CNV events. In histograms those are overplotted on the standard color scheme - e.g. red areas where high level plots were detected, by their absolute frequency. In the example plot above we performed a search for high-level gains involving EGFR, in glioblastomas, and then only plotted chromosomes 7 and 9. While the ~100% peak at EGFR is expected, additionally nearly 70% of the matched samples have a focal and high level deletion involving the CDKN2A locus.
Continue readingcancercelllines.org listed in Expasy
Entry in the Swiss Institute of Bioinformatics Catalogue
 Our recently launched cancer cell line genomics site cancercelllines.org is now listed as one of the resources in the Swiss Institute of Bioinformatics’ Expasy catalogue.
Our recently launched cancer cell line genomics site cancercelllines.org is now listed as one of the resources in the Swiss Institute of Bioinformatics’ Expasy catalogue.
cancercelllines.org - a Novel Resource for Genomic Variants in Cancer Cell Lines
DATABASE Article
Rahel Paloots and Michael Baudis¶
Database (Oxford). 2024 Apr 30:2024:baae030. doi: 10.1093/database/baae030¶
bioarXiv preprint (2023-12-13): https://doi.org/10.1101/2023.12.12.571281¶
 Abstract: Cancer cell lines are an important component in biological and medical research, enabling studies of cellular mechanisms as well as the development and testing of pharmaceuticals. Genomic alterations in cancer cell lines are widely studied as models for oncogenetic events and are represented in a wide range of primary resources. We have created a comprehensive, curated knowledge resource - cancercelllines.org - with the aim to enable easy access to genomic profiling data in cancer cell lines, curated from a variety of resources and integrating both copy number and single nucleotide variants (SNVs) data. We have gathered over 5,600 copy number profiles as well as SNV annotations for 16,000 cell lines and provide this data with mappings to the GRCh38 reference genome. Both genomic variations and associated curated metadata can be queried through the GA4GH Beacon v2 API and a graphical user interface with extensive data retrieval enabled using GA4GH data schemas under a permissive licensing scheme.
Abstract: Cancer cell lines are an important component in biological and medical research, enabling studies of cellular mechanisms as well as the development and testing of pharmaceuticals. Genomic alterations in cancer cell lines are widely studied as models for oncogenetic events and are represented in a wide range of primary resources. We have created a comprehensive, curated knowledge resource - cancercelllines.org - with the aim to enable easy access to genomic profiling data in cancer cell lines, curated from a variety of resources and integrating both copy number and single nucleotide variants (SNVs) data. We have gathered over 5,600 copy number profiles as well as SNV annotations for 16,000 cell lines and provide this data with mappings to the GRCh38 reference genome. Both genomic variations and associated curated metadata can be queried through the GA4GH Beacon v2 API and a graphical user interface with extensive data retrieval enabled using GA4GH data schemas under a permissive licensing scheme.
Availability and Implementation: Our resource is publicly available on the web at cancercelllines.org.
Continue readingProgenetix as SIB and ELIXIR Resource
Recognizing the Progenetix platform as Swiss contribution to the European bioinformatics resources ecosystem
 The Progenetix resource has finally been recognized as an official contribution to the ELIXIR European bioinformatics informatics ecosystem. Besides Expasy Progenetix now is linked through ELIXIR's resource page. Or just go directly to progenetix.org (and its daughter project cancercelllines.org).
The Progenetix resource has finally been recognized as an official contribution to the ELIXIR European bioinformatics informatics ecosystem. Besides Expasy Progenetix now is linked through ELIXIR's resource page. Or just go directly to progenetix.org (and its daughter project cancercelllines.org).
Implementing alphanumeric filters
Age queries as first use case for comparator queries
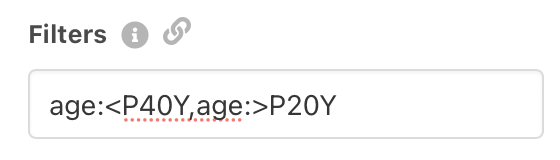 While the Beacon v2 API has in principle support for an
While the Beacon v2 API has in principle support for an alphanumeric filter
type so far in bycon
there had been no dedicated support. With yesterday's v1.0.49 update the library
now supports such queries, implemented & tested specifically for age (...at diagnosis)
values.
Plotting now handled by Python - Goodbye Perl and PGX
The plotting code has been re-implemented as part of the bycon
The last month has seen the transition from the Perl-based PGX plotting apps
to new libraries implemented as part of the main bycon
Python library stack. Very few of the options have been removed although some
(probably color side bars for clustering items ...) will be added again.
New plotting documentation and changes in parameter handling
Separate page for plot options and parameters
Inspired by some request regarding plotting of large sample numbers we've added
a dedicated page to this site as the one-stop place for plot generation
information (though you still will find examples e.g. on the use-cases page).
Plot parameters lost the - prefix
We have removed the - prefix from the plot parameter names; e.g. the previous
-plotChros selector is now simply plotChros.
New .pgxfreq Interval Frequencies File Type
Changed suffix and upcoming additions...
With the November 2022 update we changed the file suffix to pgxfreq to keep a
clean separation between the (usually binned) CNV frequency files and the
(usually raw) representation of sample-specific CNVs (and other variants).
Geographic Maps
Displaying geolocations query results or user-provided data on a map
This new feature utilizes the geolocations service to:
- display of matched cities on a map using the
&output=mapoption - load arbitrary data from a hosted data table (e.g. on Github)
Variant Types Update
Correcting Hierarchical Queries for Variant Type
The variantType query parameter - recommended only for non-precise variants, i.e. such w/o a
specified allele - is now being expanded correctly. In Progenetix these are only CNVs, all
expressed as (sub)classes of EFO:0030066 (relative copy number variation):
EFO:0030066:
child_terms:
- EFO:0030066
- EFO:0030067
- EFO:0030068
- EFO:0030069
- EFO:0030070
- EFO:0030071
- EFO:0030072
- EFO:0030073
EFO:0030070:
child_terms:
- EFO:0030070
- EFO:0030071
- EFO:0030072
- EFO:0030073
...etc. Continue reading
Implementation of the GA4GH Beacon protocol for discovery and sharing of genomic copy number variation data
Poster Abstract | ESHG Vienna 2022
Background & Objectives Genomic copy number variations (CNV) are a major contributor to inter-individual genomic variation, can be causative events in rare diseases, but especially represent the majority of the mutational landscape in the most malignancies. While specific CNV events and some recurring patterns have contributed to the identification of individual cancer drivers and the recognition of cancer subtypes, the complexity of genomic CNV patterns requires large amounts of well-defined genomic profiles for statistically meaningful analyses. At the other end of the spectrum, in the area of rare disease genomics the potential pathogenicity of individual CNV events requires validation against a vast set of disease-related and reference genomic profiles and annotations.
Continue readingCNVs in Prenatal Tests & Maternal Malignancies
Publication indicating rare CNV signatures from a nationwide Dutch screening program
In a new publication in the Journal of Clinical Oncology CJ Heesterbeek, SM Aukema and the co-authors from the Dutch NIPT Consortium report about the incidence and diagnostic significance of incidential detection om maternal copy number variations in a large screening program aimed at detecting chromosomal imbalances in embryos, for a prediction of developmental abnormalities.
Continue readingPaginated Downloads
Chunk-wise downloads of search results
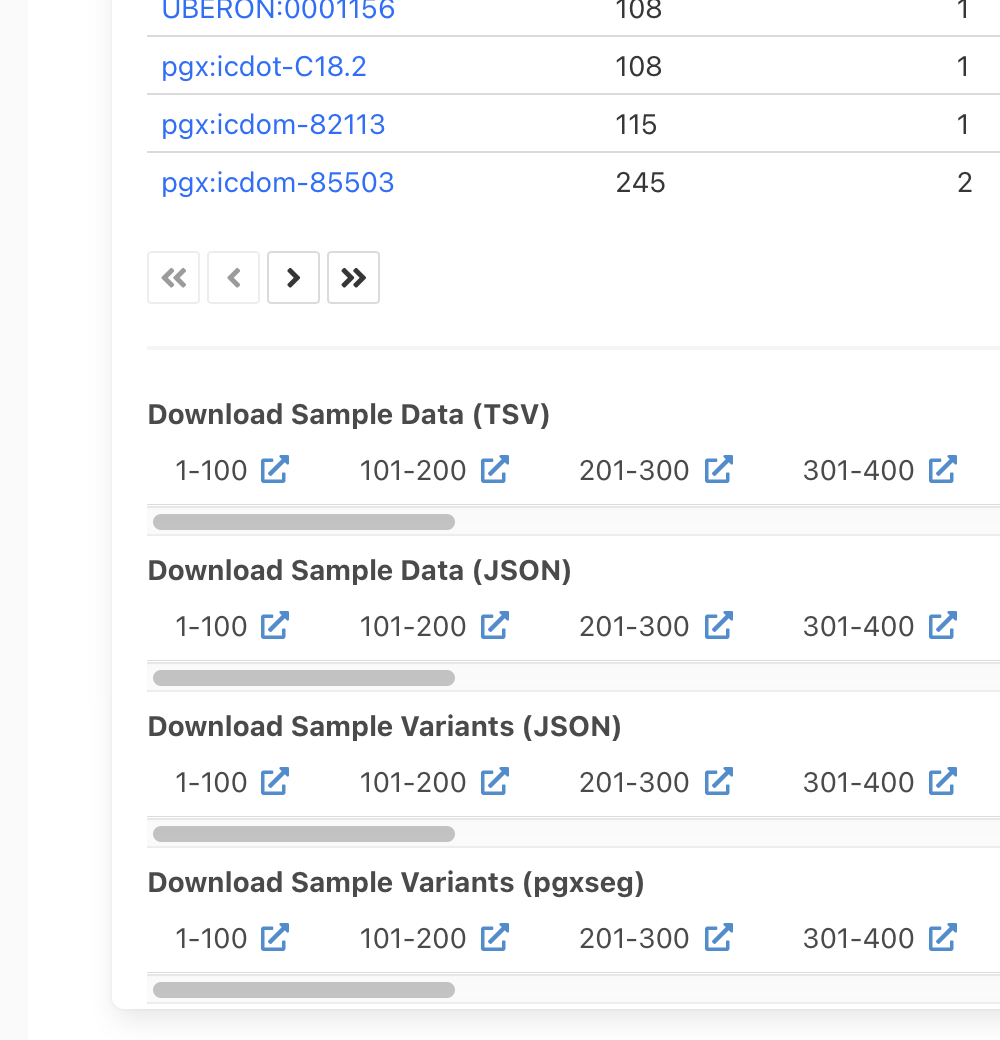 Throught its Search Samples page Progenetix has
always offered options to download search results (biosamples, variants) in different formats (JSON,
tab-delimited tables, pgxseg files ...). However, especially for large results with
thousands of samples and potentially millions of variants this led to inconsistent behaviour
e.g. time-outs or dropped connections.
Throught its Search Samples page Progenetix has
always offered options to download search results (biosamples, variants) in different formats (JSON,
tab-delimited tables, pgxseg files ...). However, especially for large results with
thousands of samples and potentially millions of variants this led to inconsistent behaviour
e.g. time-outs or dropped connections.
Now, API responses are capped through the limit parameter to default "sensible" values
which, however, can be adjusted for systematic data access & retrieval. This functionality
is also implemented in the sample search form, allowing e.g. the limited retrieval of
a subset of samples from large or general cancer types, or the "paging" through consecutive
sample groups for partitioned data retrieval.
VRSified Variants
Variant Response in GA4GH Variant Representation Standard (VRS) Format
The variant format served through the API has now changed to a format commpatible with the GA4GH Variant Representation Standard (VRS version (bleeding edge version...).
json
{
"caseLevelData": [
{
"analysisId": "pgxcs-kftwfurn",
"biosampleId": "pgxbs-kftvj7rz",
"id": "pgxvar-5c86664409d374f2dc4eeb93"
}
],
"variation": {
"location": {
"interval": {
"end": {
"type": "Number",
"value": 62947165
},
"start": {
"type": "Number",
"value": 23029501
}
},
"sequenceId": "refseq:NC_000018.10",
"type": "SequenceLocation"
},
"relativeCopyClass": "partial loss",
"updated": "2022-03-29T15:06:46.526020",
"variantInternalId": "18:23029501-62947165:DEL"
}
}
json
{
"caseLevelData": [
{
"analysisId": "pgxcs-kftwfurn",
"biosampleId": "pgxbs-kftvj7rz",
"id": "pgxvar-5c86664409d374f2dc4eeb93"
}
],
"variantInternalId": "18:23029501-62947165:DEL"
"referenceName": "18",
"start": 23029501,
"end": 62947165,
"variantType": "DEL",
"updated": "2022-03-29T15:06:46.526020"
}
json
{
"caseLevelData": [
{
"analysisId": "pgxcs-kl8hg4ky",
"biosampleId": "pgxbs-kl8hg4ku",
"id": "pgxvar-5be1840772798347f0eda0d8"
}
],
"variation": {
"location": {
"interval": {
"end": {
"type": "Number",
"value": 7577121
},
"start": {
"type": "Number",
"value": 7577120
},
"type": "SequenceInterval"
},
"sequenceId": "refseq:NC_000017.11",
"type": "SequenceLocation"
},
"state": {
"sequence": "G",
"type": "LiteralSequenceExpression"
},
"updated": "2022-03-29T15:35:35.700954",
"variantInternalId": "17:7577121:C>G"
}
}
json
{
"caseLevelData": [
{
"analysisId": "pgxcs-kl8hg4ky",
"biosampleId": "pgxbs-kl8hg4ku",
"id": "pgxvar-5be1840772798347f0eda0d8"
}
],
"variantInternalId": "17:7577121:C>G",
"start": 7577120,
"end": 7577121,
"referenceName": "17",
"referenceBases": "C",
"alternateBases": "G",
"updated": "2022-03-29T15:35:35.700954"
}
Histogram Improvements
Excluding reference samples from default plots
So far all samples matching a grouping code ("collation"; disease, publication etc.)
have been included when generating the pre-computed CNV frequencies. However, the
potential inclusion of normal/refernce samples sometimes lead to "dampened" CNV
profiles. Now, samples labeled as "reference sample" (EFO:0009654) -
a term we had introduced into the Experimental Factor Ontology - are excluded from
pre-computed histograms. However, when e.g. calling up samples from publications
using the search panel referencve samples will be included unless specifically excluded.
Query-based histograms
Direct generation of histogram plots from Beacon queries
So far, the plot API only provided (documented) access to generate CNV histogram
plots from "collations" with pre-computed frequencies.
The bycon API now offers a direct access to the histograms, by adding &output=histoplot
to a Beacon (biosamples) query URL. The server will first query the samples and then perform
a handover to the plotting API. Please be aware that this procedure is best suited for limited
queries and may lead to a time-out.
Genomic Interval Changes
New positions for the 1Mb interval maps
So far, CNV histograms and .pgxseg segment and matrix files used a 1Mb genome binning, based on the consecutive assignment of 1Mb intervals from 1pter -> Yqter. This resulted in 3102 intervals, with the last interval of each chromosome being smaller.
On 2022-02-11 we have changed the procedure. Now, the last interval of the short arm of any chromosome is terminated at the centromere, leading to
- a (potentially) shortened "last p" interval
- a shift of most interval positions
- a changed interval number from 3102 to 3106
CNV Ontology Proposal - Now Live at EFO
EFO Ontology contains now terms for (relative) CNV levels
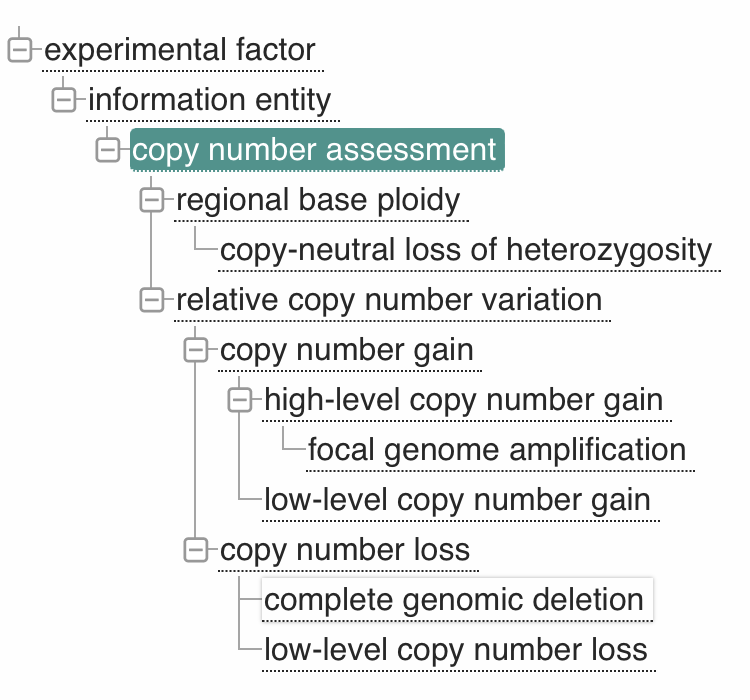 As part of the hCNV-X work - related to "Workflows and Tools for hCNV Data Exchange
Procedures" and to the intersection with Beacon and GA4GH VRS - we have now a new
proposal
for the creation of an ontology for the annotation of (relative) CNV events. The CNV
representation ontology is targeted for adoption by Sequence Ontology (SO)
and then to be used by an updated version of the VRS standard. Please see the
discussions linked from the proposal page.
However, we have also contributed the CNV proposal to EFO where it has gotten
live on January 21.
As part of the hCNV-X work - related to "Workflows and Tools for hCNV Data Exchange
Procedures" and to the intersection with Beacon and GA4GH VRS - we have now a new
proposal
for the creation of an ontology for the annotation of (relative) CNV events. The CNV
representation ontology is targeted for adoption by Sequence Ontology (SO)
and then to be used by an updated version of the VRS standard. Please see the
discussions linked from the proposal page.
However, we have also contributed the CNV proposal to EFO where it has gotten
live on January 21.
Introducing variant_state classes for CNVs
More granular annotation of CNV types
More information can be found in the description of ontology use for CNVs.
Continue readingTerm-specific queries
Allowing the de-selection of descendant terms in ontology filters
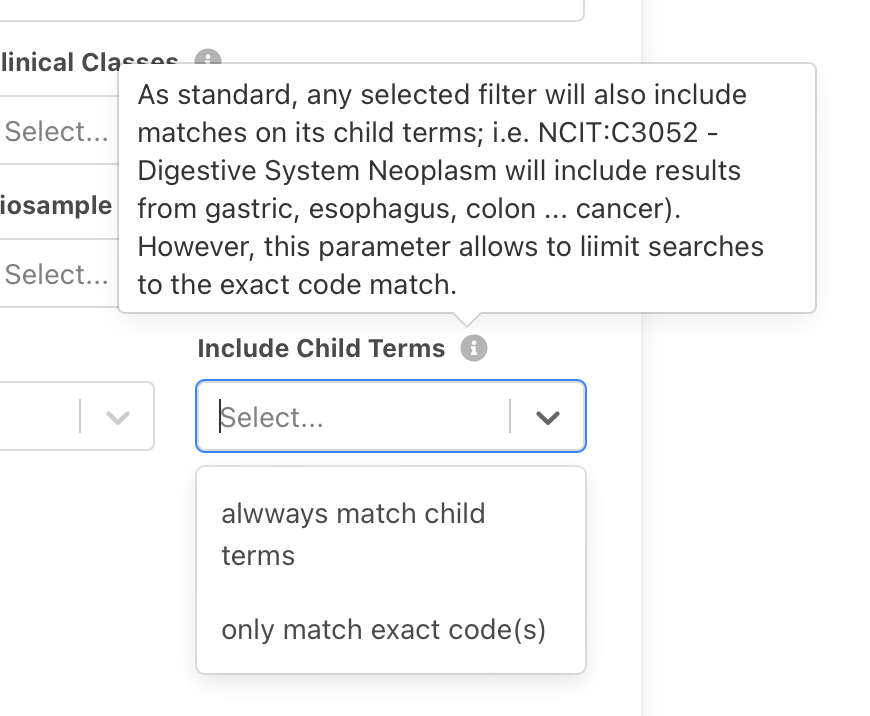 So far (and still as standard), any
selected filter will also include matches on its child terms; i.e. "NCIT:C3052 -
Digestive System Neoplasm" will include results from gastric, esophagus, colon
... cancer. Here we introduce a selector for the search panel to make use of the Beacon v2
filters
So far (and still as standard), any
selected filter will also include matches on its child terms; i.e. "NCIT:C3052 -
Digestive System Neoplasm" will include results from gastric, esophagus, colon
... cancer. Here we introduce a selector for the search panel to make use of the Beacon v2
filters includeDescendantTerms pragma, which can be set to false if one only
wants to query for the term itself and exclude any child terms from the matching.
Please be aware that this can only be applied globally and will affect all filtering terms used in a query. More information is available in the Filtering Terms documentation.
Continue readingBUG FIX Frequency Maps
Fix "only direct code matches" frequencies
Pre-computed Progenetix CNV frequency histograms (e.g. for NCIT codes) are based
samples from all child terms; e.g. NCIT:C3262 will display an overview of all
neoplasias, although no single case has this specific code.
However, there had been a bug when under specific circumstances (code has some mapped samples and code has more samples in child terms) only the direct matches were used to compute the frequencies although the full number of samples was indicated in the plot legend. FIXED.
Continue readingPublications - Updated publication listings
Progenetix citations page and better map
Progenetixuse Page¶
We have introduced a new publications listing page which contains links to articles that cite or use Progenetix and resources from this "ecosystem." Please let us know if you are aware of other such cases - frequently the publications do not use a proper citation format but just refer to "according tho the Progenetix resource" or similar in the text.
Continue readingAPI: Biosample Schema Update
Conversion of `biocharacteristics` array to separate parameters
The Biosample schema used for exporting Progenetix data has been adjusted with respect to representation of "bio-"classifications. The previous biocharacteristics list parameter has been deprecated and its previous content is now expressed in1:
histologicalDiagnosis(PXF)sampledTissue(PXF)icdoMorphology(pgx)icdoTopography(pgx)
Progenetix - An open reference resource for copy number vatiation data in cancer
Qingyao Huang¶
Cancer Genomics Consortium Annual Meeting 2021 Aug 1-4¶

Additional Links¶
Continue readingAPI: Beacon Paths Updates
For testing the rapidly evolving Beacon v2 API, we have now implemented more paths/endpoints which mostly conform to the brand new & still "flexible" v2.0.0-draft.4 version. Please check the documentation and examples.
Continue readingAPI: JSON Exports now camelCased
In "forward-looking" conformity with the Beacon v2
API, the JSON attributes of the API responses has been changed from snake_cased
to camelCased. Please adjust your code, where necessary.
The Progenetix oncogenomic resource in 2021
The Progenetix oncogenomic resource in 2021¶
Qingyao Huang, Paula Carrio Cordo, Bo Gao, Rahel Paloots, Michael Baudis¶
Database (Oxford). 2021 Jul 17;2021:baab043.¶
- doi: 10.1093/database/baab043.
- pubmed: 34272855
- PMCID: PMC8285936.
- bioRxiv. doi: doi.org/10.1101/2021.02.15.428237
 This article provides an overview of recent changes and additions to the Progenetix database and the services provided through the resource.
This article provides an overview of recent changes and additions to the Progenetix database and the services provided through the resource.
New feature - LOH data
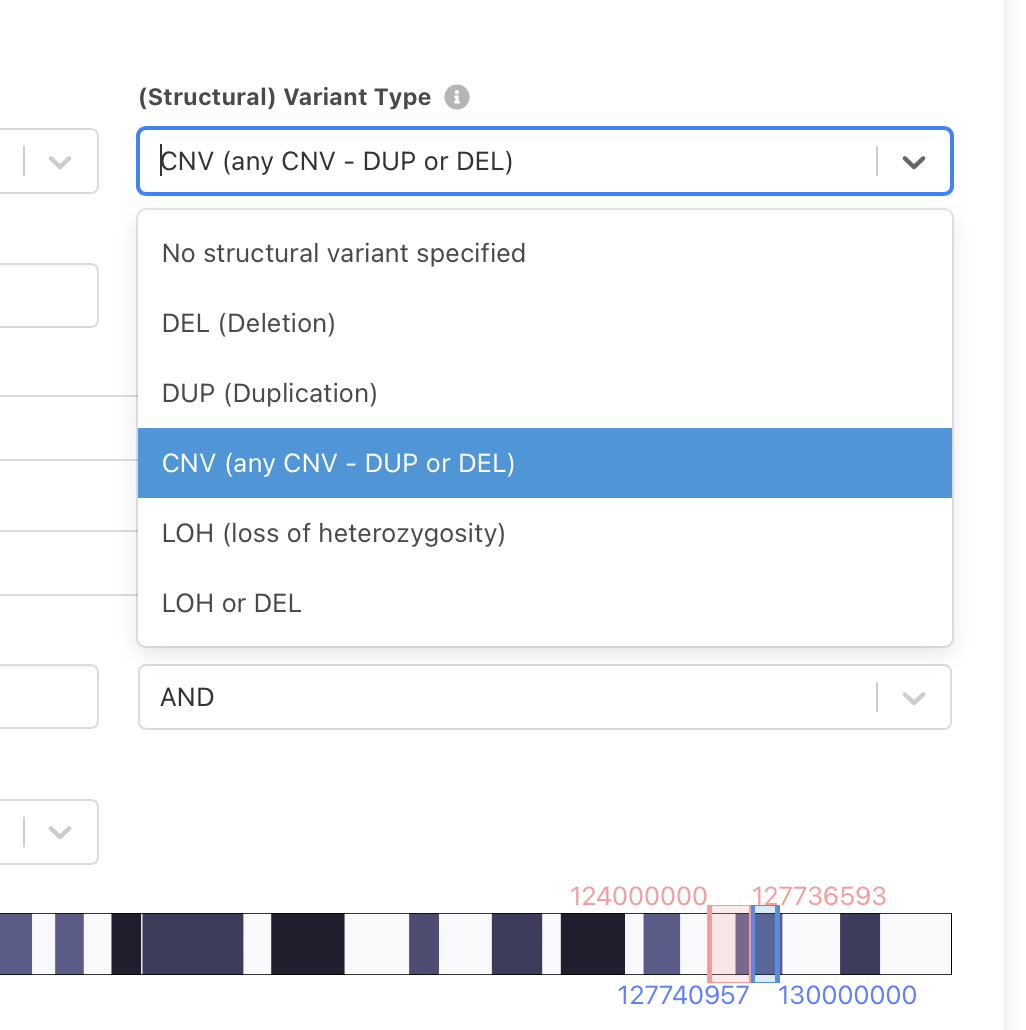 Loss of heterozygosity (LOH) is a phenomenon frequently observed in cancer genomes where the selective pressure to keep only the susceptible gene product from one allele removes the other healthy allele from the pool; In this context, copy neutral - loss of heterozygosity (CN-LOH) is commonly observed in haematological malignancies (O'keefe et al., 2010 and Mulligan et al., 2007). To Progenetix oncogenomic resource, comprising of nearly 800 cancer types (by NCIt classification) as of 2021, we expanded the new feature of LOH in our data collection, in addition to the total copy number, to open the door for the analysis of frequency and impact of this phenomenon.
Loss of heterozygosity (LOH) is a phenomenon frequently observed in cancer genomes where the selective pressure to keep only the susceptible gene product from one allele removes the other healthy allele from the pool; In this context, copy neutral - loss of heterozygosity (CN-LOH) is commonly observed in haematological malignancies (O'keefe et al., 2010 and Mulligan et al., 2007). To Progenetix oncogenomic resource, comprising of nearly 800 cancer types (by NCIt classification) as of 2021, we expanded the new feature of LOH in our data collection, in addition to the total copy number, to open the door for the analysis of frequency and impact of this phenomenon.
Update 2021-01-28:
LOH variants can now be queried through the Search and Beacon+ interfaces, either as specific variants or together with deletions.
Please be aware that in contrast to the "complete for chromosomes 1-22" DUP and DEL calls, LOH is only determined for a subset of samples and therefore will be underreported in the statistics section.
Continue readingImproved Data Access through Histograms
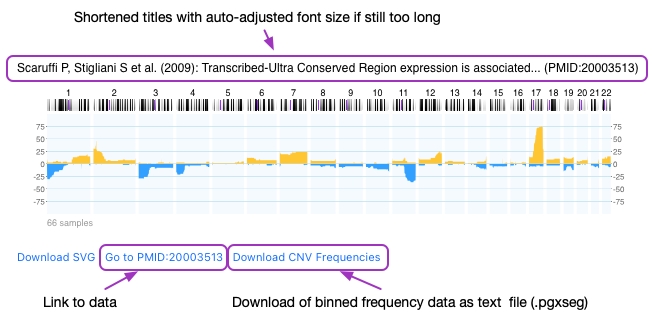
Histograms for static datasets (e.g. NCIT codes, publications ...) provide now links to the dataset details page as well as a download option for the binned CNV frequency data.
Continue readingSignatures of Discriminative CNA in 31 Cancer Subtypes
Bo Gao and Michael Baudis (2021)¶
Accepted at Frontiers in Genetics, 2021-04-15¶
Abstract¶
Copy number aberrations (CNA) are one of the most important classes of genomic mutations relatedto oncogenetic effects. In the past three decades, a vast amount of CNA data has been generated bymolecular-cytogenetic and genome sequencing based methods. While this data has been instrumentalin the identification of cancer-related genes and promoted research into the relation between CNA andhisto-pathologically defined cancer types, the heterogeneity of source data and derived CNV profilespose great challenges for data integration and comparative analysis. Furthermore, a majority of exist-ing studies have been focused on the association of CNA to pre-selected ”driver” genes with limitedapplication to rare drivers and other genomic elements.
Continue readingBeacon+ and Progenetix Queries by Gene Symbol
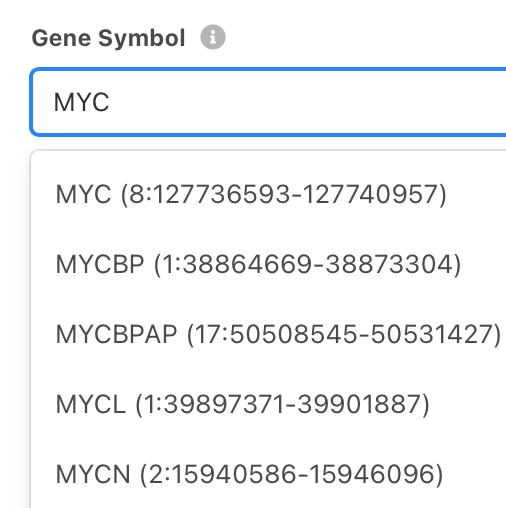 We have introduced a simple option to search directly by Gene Symbol, which will match to any genomic variant with partial overlap to the specified gene. This works by expanding the Gene Symbol (e.g. TP53, CDKN2A ...) into a range query for its genomic coordinates (maximum CDR).
We have introduced a simple option to search directly by Gene Symbol, which will match to any genomic variant with partial overlap to the specified gene. This works by expanding the Gene Symbol (e.g. TP53, CDKN2A ...) into a range query for its genomic coordinates (maximum CDR).
Such queries - which would e.g. return all whole-chromosome CNV events covering the gene of interest, too - should be narrowed by providing e.g. Variant Type and Maximum Size (e.g. 2000000) values.
Diffuse Intrinsic Pontine Glioma (DIPG) cohort
Diffuse Intrinsic Pontine Glioma (DIPG) is a highly aggressive tumor type that originate from glial cells in the pon area of brainstem, which controls vital functions including breathing, blood pressure and heart rate. DIPG occurs frequently in the early childhood and has a 5-year survival rate below 1 percent. Progenetix has now incorporated the DIPG cohort, consisting of 1067 individuals from 18 publications. The measured data include copy number variation as well as (in part) point mutations on relevant genes, e.g. TP53, NF1, ATRX, TERT promoter.
Continue readingTCGA CNV Data
 www.cancer.gov/tcga) for quite some time, we have now launched a dedicated search page to facilitate data access and visualization using the standard Progenetix tools.
www.cancer.gov/tcga) for quite some time, we have now launched a dedicated search page to facilitate data access and visualization using the standard Progenetix tools.
Additionally, the TCGA page section priovides pre-computed CNV frequency data for the individual TCGA studies.
Continue readingarrayMap is Back
 After some months of dormancy, the arrayMap resource has been relaunched through integration with the new Progenetix site. All of the oiginal arrayMap data has now been integrated into Progenetix, and of today the
After some months of dormancy, the arrayMap resource has been relaunched through integration with the new Progenetix site. All of the oiginal arrayMap data has now been integrated into Progenetix, and of today the arraymap.org domain maps to a standard Progenetix search page, where only data samples with existing source data (e.g. probe specific array files) will be presented.
Website updates
The new year brings some refinements to biosamples search and display:
- added example for a pure filter search (HeLa)
- made UCSC link depending on variants
- added info pop-ups to biosamples table header
- removed DEL and DUP fractions from biosamples table
- added label display for
external_referencesitems in biosamples table
Enjoy ...
Continue readingGenomic Copy Number Signatures...
Genomic Copy Number Signatures Based Classifiers for Subtype Identification in Cancer¶
Bo Gao and Michael Baudis (2020)¶
bioRxiv, 2020-12-18¶
Continue readingAPI and Services Documentation
Following the launch of the updated Progenetix website (new interface, now much
more data with >130'000 samples...) and the recent introduction of the new
Python based bycon API for BeaconPlus and Progenetix Services
we now also have some structured information for the different API options.
pgx namespace and persistant identifiers
While the pgx prefix had been registered in 2017 with identifiers.org
we recently changed the resolver and target mappings on the Progenetix server.
This went hand-in-hand with the generation of unique & persistant identifiers
for the main data items.
bycon powered BeaconPlus
Moving to a new, Python-based API
We've changed the Beacon backend to the bycon code base. The new project's
codebase is accessible through the bycon
project. Contributions welcome!
Example¶
Continue readingGA4GH Beacon v2 at GA4GH Plenary
GA4GH Beacon v2 - Evolving Reference Standard for Genomic Data Exchange¶
GA4GH 8th Plenary¶
Gary Saunders, Jordi Rambla de Argila, Anthony Brookes, Juha Törnroos and Michael Baudis¶
For the ELIXIR Beacon project, GA4GH Discovery work stream and the international network of Beacon API developers¶
The Beacon driver project was one of the earliest initiatives of the Global Alliance for Genomics and Health with the Beacon v1.0 API as first approved GA4GH standard. Version 2 of the protocol is slated to provide fundamental changes, towards a Internet of Genomics foundational standard:
Continue readingProgenetix at GA4GH 2020 Plenary
GA4GH 8th Plenary¶
Michael Baudis¶
The Progenetix oncogenomics resource provides sample-specific cancer genome profiling data and biomedical annotations as well as provenance data from cancer studies. Especially through currently 113322 curated genomic copy number number (CNV) profiles from 1600 individual studies representing over 500 cancer types (NCIt), Progenetix empowers aggregate and comparative analyses which vastly exceed individual studies or single diagnostic concepts.
Continue readingNew Progenetix Website
The Progenetix website has been completely rebuilt using a JavaScript / React based framework and API based content delivery. At its core, the site is built around the Beacon standard, with some extensions for data colections and advanced query options.
Continue readingProgenetix now licensed under CC-BY 4.0
After many years of using a CreativeCommons CC-BY-SA ("attribution + share alike"), the Progenetix resource has dropped the "SA - share alike" attribute and is now "attribution" only. This may facilitate the use of the data in more complex and/or commercial scenarios - enjoy!
Continue readingError Calibration ... for CNA Analysis
Minimum Error Calibration and Normalization for Genomic Copy Number Analysis.¶
Bo Gao and Michael Baudis (2020)¶
bioRxiv, 2019-07-31. DOI 10.1101/720854¶
Genomics, Volume 112, Issue 5, September 2020, Pages 3331-3341, accepted 2020-05-06 doi.org/10.1016/j.ygeno.2020.05.008.¶
Continue readingGeographic assessment of cancer genome profiling studies
Geographic assessment of cancer genome profiling studies.¶
Paula Carrio Cordo, Elise Acheson, Qingyao Huang and Michael Baudis (2020)¶
DATABASE, Volume 2020, 2020, baaa009, doi.org/10.1093/database/baaa009¶
Continue readingCURIE Prefix Remapping - NCIT & pubmed
Wherever possible, data annotation in Progenetix uses {S}[B] OntologyClass
objects for categorical values, with CURIEs as id values. So far, the
Progenetix databases had used pubmed: for PubMed identifiers and ncit:
for NCI Metathesaurus (Neoplasm) ids.
Population assignment from cancer genomes
Enabling population assignment from cancer genomes with SNP2pop.¶
Huang Q and Baudis M. (2020)¶
Sci Rep 10, 4846 (2020). doi.org/10.1038/s41598-020-61854-x¶
Continue readingBeaconPlus in ELIXIR Beacon Network
The Beacon+ implementation of the GA4GH Beacon protocol has become a part of the ELIXIR Beacon Network, an expanding Beacon service to query multiple Beacon resources and aggregate their query results.
Continue readingBeacon Variants in UCSC Browser
The response element of the Beacon+ interface now contains a link for displaying the matched variants e.g. of e.g. a CNV query in the UCSC genome browser.
Continue readingMinimum Error Calibration and Normalization for Genomic Copy Number Analysis
Minimum Error Calibration and Normalization for Genomic Copy Number Analysis.¶
Bo Gao and Michael Baudis (2019)¶
bioRxiv, 2019-07-31. DOI 10.1101/720854¶
Continue readingNew info.progenetix.org site
Launch of new info.progenetix.org resource site¶
Today, we started to provide a new documentation structure for our group's work and software projects.
The site is assumed, over time, to replace the previous Progenetix guide.
Continue readingarrayMap 2014: an...
arrayMap 2014: an updated cancer genome resource.¶
Cai H, Gupta S, Rath P, Ai N, Baudis M.¶
Abstract Somatic copy number aberrations (CNA) represent a mutation type encountered in the majority of cancer genomes. Here, we present the 2014 edition of arrayMap (www.arraymap.org), a publicly accessible collection of pre-processed oncogenomic array data sets and CNA profiles, representing a vast range of human malignancies. Since the initial release, we have enhanced this resource both in content and especially with regard to data mining support.
Continue readingChromothripsis-like...
Chromothripsis-like patterns are recurring but heterogeneously distributed features in a survey of 22,347 cancer genome screens.¶
Cai H, Kumar N, Bagheri HC, von Mering C, Robinson MD, Baudis M.¶
Abstract BACKGROUND: Chromothripsis is a recently discovered phenomenon of genomic rearrangement, possibly arising during a single genome-shattering event. This could provide an alternative paradigm in cancer development, replacing the gradual accumulation of genomic changes with a "one-off" catastrophic event. However, the term has been used with varying operational definitions, with the minimal consensus being a large number of locally clustered copy number aberrations.
Continue readingProgenetix: 12 years...
Progenetix: 12 years of oncogenomic data curation.¶
Cai H, Kumar N, Ai N, Gupta S, Rath P, Baudis M.¶
Abstract DNA copy number aberrations (CNAs) can be found in the majority of cancer genomes and are crucial for understanding the potential mechanisms underlying tumor initiation and progression. Since the first release in 2001, the Progenetix project (www.progenetix.org) has provided a reference resource dedicated to provide the most comprehensive collection of genome-wide CNA profiles.
Continue readingProgenetix & arrayMap Changes (2012-06-01 - 2013-05-22)
2013-05-22¶
- bug fix: fixing lack of clustering for CNA frequency profiles in the analysis section
- removed "Series Search" from the arrayMap side bar; kind of confusing - just search for the samples & select the series
2013-05-12¶
- introduced a method to combine sample annotations and segmentation files for user data processing (see "FAQ & GUIDE")
- fixed some array plot presentation and replotting problems
2013-05-05¶
- consolidation of script names - again, don't use deep links (besides for "api.cgi?...")
- moving of remaining sample selection options (random sample number, segments number, age range) to the sample selection page, leaving the pre-analysis page (now "prepare.cgi") for plotting/grouping options
- fixed the KM-style survival plots
2013-04-10¶
- re-factoring of the cytobands plotting for histograms and heatmaps; this also fixes missing histogram tiles
- analysis output page: the circular histogram/connections plot and group specific histograms are now all available as both SVG and PNG image files
2013-04-06¶
Some changes to the plotting options:
- the circular plot is now added as a default; and connections are drawn in for <= 30 samples (subject to change)
- one can now mark up multiple genes (or other loci of interest), for all plot types
2013-03-25¶
- added option to create custom analysis groups based on text match values
- rewritten circular plot code
2013-02-27¶
- copied data for PMIDs 17327916, 17311676, 18506749 and 18246049 from arrayMap to Progenetix
2013-02-24¶
- bug fix: gene selector was broken for about a week; fixed
2013-02-17¶
- In many places, images are now converted sever side to PNG data streams and embedded into the web pages. This will substantially decrease web data traffic and page download times. Fully linked SVG images (including region links etc.) are still available through the analysis pipeline.
2013-02-13¶
- data fix: pubmed 18160781 had missing loss values (due to irregular character encoding); fixed, thanks to Emanuela Felley-Bosco for the note!
2012-12-14¶
- moved the region filter from the analysis to the sample selection page
- added a "mark region" option to the analysis page: one now can highlight a genome region in histograms and matrix plots
2012-11-29¶
- added "select all" option to entity lists
- implemented first version of sample-to-entity match score
- added single sample annotation input field to "User File Processing"; i.e. one can now type in CNA data for a single case, and have this visualised and similar cases listed
- added per sample CNA visualisation to the samples details listings (currently if up to 100 samples)
- added direct access to sample details listing to the subsets pages
2012-11-09¶
- adding of abstract search to the publication search page
2012-10-25¶
- introduction of a matching function for similar cases by CNA profile, accessible through the sample details pages of both Progenetix and arraymap
2012-10-22¶
- Introduction of SEER groups
2012-09-26¶
The database now contains the copy number status for different interval sizes (e.g. 1MB). With this, users can now create their own data plots (histograms etc.) using more than 10000 cancer copy number profiles with a high resolution. The options here are still being tested and improved - comments welcome!
2012-09-18¶
- added a new export file format "ANNOTATED SEGMENTS FILE", which uses the first columns for standard segment annotation, followed by some diagnostic and clinical data; i.e., the information for a case is repeated for each segment:
GSM255090 22 25063244 25193559 1 NA C50 8500/3 breast Infiltrating duct carcinoma, NOS Carcinomas: breast ca. NA 1 51 0.58
GSM255090 22 25368299 48899534 -1 NA C50 8500/3 breast Infiltrating duct carcinoma, NOS Carcinomas: breast ca. NA 1 51 0.58
GSM255091 1 2224111 30146401 -1 NA C50 8500/3 breast Infiltrating duct carcinoma, NOS Carcinomas: breast ca. NA 0 72 0.54
GSM255091 1 35418712 37555461 1 NA C50 8500/3 breast Infiltrating duct carcinoma, NOS Carcinomas: breast ca. NA 0 72 0.54
2012-09-13¶
- added gene selection for region specific replotting of array data
2012-08-22¶
- the gene database has been changed to the last version of the complete (HUGO names only) Ensembl gene list for HG18; previously, only a subset of "cancer related genes" was offered in the gene selection search fields
2012-07-04¶
- some interface and form elements have been streamlined (e.g. less commonly used selector fields, sample selection options)
- some common options are now displayed only if activated (e.g. "mouse over" to see all files available for download)
- icon quality has been enhanced for all but the details pages
2012-06-13¶
- New: All pre-generated histogram and ideogram plots are now produced based on a 1Mb matrix, with a 500Kb minimum size filter to remove CNV/platform dependent background from some high resolution array platforms. The unfiltered data can still be visualized through the standard analysis procedures.
- Bug fix: Interactive segment size filtering so far only worked for region specific queries, but not as a general filter (see above). This has been fixed; a minimum segment size in the visualization options now will remove all smaller segments.
2012-06-01¶
- NEW: change log; that is what is shown here
- FEATURE: The interval selector now has options to include the p-arms of acrocentric chromosomes (though the data itself there may be incompletely annotated!). Feature requested by Melody Lam.
arrayMap feature update(s)
arrayMap feature update(s)¶
Over the last weeks, we have introduced a number of new search/ordering features to arrayMap. Some of those mimic functions previously implemented in Progenetix. Overall, the highlights are:
 ICD entity aggregation
ICD entity aggregation- all ICD-O entities with their according samples
 ICD locus aggregation
ICD locus aggregation- all tumor loci with their according samples
 Clinical group aggregation
Clinical group aggregation- clinical super-entities (e.g. "breast ca.": all carcinoma types with locus breast) with their samples
 Publication aggregation
Publication aggregation- all publication with samples in arrayMap
In contrast to Progenetix, we do not offer precomputed SCNA histograms. However, users can generate them on the fly, but should consider the specific challenges in doing so (e.g. noise background in frequency calculations).
Continue readingGenomic imbalances in 5918 malignant tumors
Genomic imbalances in 5918 malignant epithelial tumors: an explorative meta-analysis of chromosomal CGH data.¶
Baudis M.¶
Abstract BACKGROUND: Chromosomal abnormalities have been associated with most human malignancies, with gains and losses on some genomic regions associated with particular entities. METHODS: Of the 15429 cases collected for the Progenetix molecular-cytogenetic database, 5918 malignant epithelial neoplasias analyzed by chromosomal Comparative Genomic Hybridization (CGH) were selected for further evaluation. For the 22 clinico-pathological entities with more than 50 cases, summary profiles for genomic imbalances were generated from case specific data and analyzed.
Continue readingOnline database and...
Online database and bioinformatics toolbox to support data mining in cancer cytogenetics.¶
Baudis M.¶
Continue readingProgenetix.net: an...
Progenetix.net: an online repository for molecular cytogenetic aberration data.¶
Baudis M, Cleary ML.¶
Abstract Through sequencing projects and, more recently, array-based expression analysis experiments, a wealth of genetic data has become accessible via online resources. In contrast, few of the (molecular-) cytogenetic aberration data collected in the last decades are available in a format suitable for data mining procedures. www.progenetix.net is a new online repository for previously published chromosomal aberration data, allowing the addition of band-specific information about chromosomal imbalances to oncologic data analysis efforts.
Continue reading